6.2 Evolution by gene duplications and altered regulatory environments
However, the analysis of paralog regulation with respect to three-dimensional genome folding allows further conclusions about the evolutionary history of genes and how new functionality by novel genes is most efficiently created during evolution. Loss-of-function mutations degrade most young duplicated genes into pseudogenes (Lynch and Conery 2003). These observations raise the question of which mechanisms led to the survival and expression of the many paralog genes present in the human genome. Some models explain the survival of paralogs by sub-functionalization, neo-functionalization or sharing of gene dosage (Innan and Kondrashov 2010). Our observation of co-regulated paralogs in the regulatory environments of TADs is consistent with a recent study of expression patterns between young and old paralogs (Lan and Pritchard 2016). This study often finds one copy of young paralogs to be downregulated, indicating that dosage-sharing allows for initial survival of duplicated genes and slow functional adaption later on (Lan and Pritchard 2016).
The creation of paralogs by tandem duplication is an efficient evolutionary mechanism to introduce additional functionality to a genome by keeping the original gene unaffected. We observe that very close paralogs seems to have decreased Hi-C contacts between their promoters when located in the same TAD, but show enriched association to the same regulatory elements. This somewhat unexpected phenomenon can be explained by a subset of paralogs that functionally replace each other, for example as alternative units in protein complexes. This replacing functionality would require actively regulated exclusive expression of paralogs. Interestingly, a similar mechanism was reported for olfactory receptor genes that contact each other even across different chromosomes to facilitate particular expression of only a single receptor gene per cell (Monahan and Lomvardas 2015; Monahan et al. 2017).
The results of our analysis together with recent literature suggest a many-step mechanism of paralog evolution (Fig. 6.2). First, tandem duplications generate paralog genes, initially coregulated within TADs. Then subsequent chromosomal rearrangements might relocate paralogs to loci distant from their homologs but retain increased chromatin contacts. Further, reorganization of the regulatory environment allows their increased independence being eventually placed even on different chromosomes where contact is no longer necessary. An exception would be genes that precise to be strongly co-regulated with the original copy, for example, to produce replacement proteins.
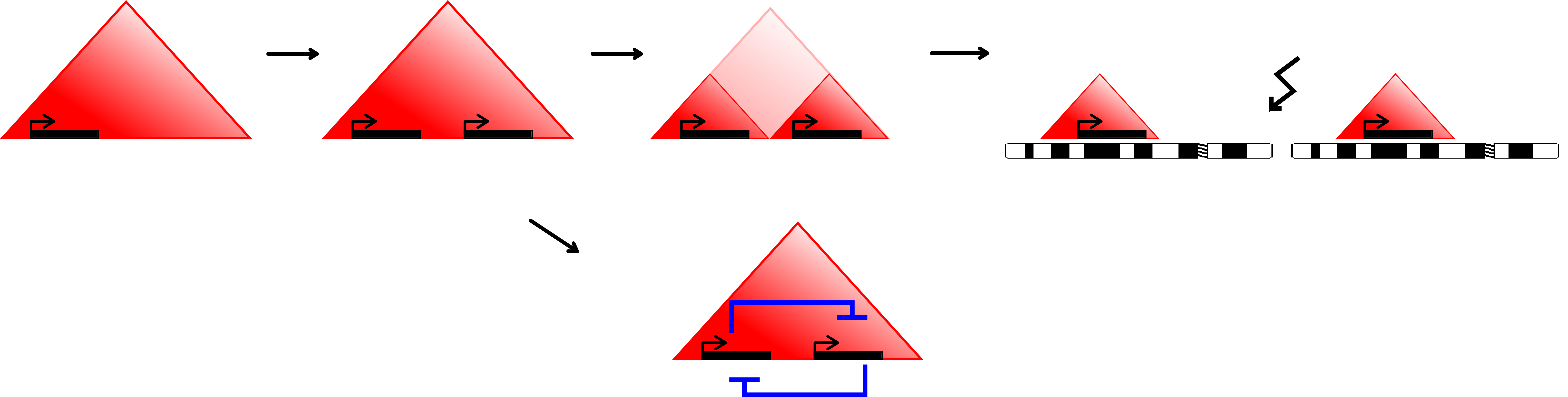
Figure 6.2: Model of paralog gene creation in TADs and evolving regulatory context and TADs over time. Tandem-duplication accounts for the creation of most paralogs. Duplication within a TAD allows the new copy of the gene to be regulated by the same environment (top left). Over evolutionary time, the interactions pattern and regulatory input might change to allow for more independent expression regulation (top center). Evolutionary rearrangements might even relocate the genes to different chromosomes (top right). Alternatively, the two copies might stay in the same TAD to enable them to be coordinately regulated (bottom).
Several mechanisms can explain the creation of new functionality and morphological diversity between species across evolution. A key component and driver for new gene function in evolution or neo-functionalization can be the birth of new enhancers through the acquisition of transcription factor binding and subsequent novel regulatory functions (Long et al. 2016). Similarly, deletion or duplication of TAD boundaries could change the folding structure of genomic regions and enable regulatory contacts with previously insulated regulatory elements (Ibn-Salem et al. 2014; Lupiáñez et al. 2015; Franke 2017). In agreement with these mechanisms observed in genetic diseases, we find that evolutionary rearrangements disrupting TADs are associated with divergent gene expression patterns between mouse and human. These findings indicate an evolutionary mechanism by which structural variations lead to altered domain organization with associated expression changes. Expression in different tissues or developmental time points might enable novel functions and morphologies. One example of altered physiological morphologies upon structural variations is the Liebenberg Syndrom that is characterized by malformation of upper limbs, showing the characteristic bone morphology of hind-limbs (Spielmann et al. 2012). This syndrome is caused by deletions that disrupt a TAD boundary element or translocations. Bothe alternations expose the PITX1 gene to enhancers with specific lower-limb activity (Spielmann and Mundlos 2013). This example, together with other pathogenic phenotypes caused by TAD disruptions (Lupiáñez et al. 2015; Franke et al. 2016; Flöttmann et al. 2017), shows that altered genome folding upon TAD disruption can potentially lead to physiological and morphological novelties during evolution.
Together, our data suggest that TADs act as protective nests for evolving newcomer genes. Duplicating genes within TADs seems to be a reasonable evolutionary mechanism, much more straightforward than creating from nothing an entirely new regulatory environment for a new gene. Genetic variations that alter genome folding and expose genes to new regulatory environments can then create new functions of the gene by expression in different contexts leading to physiological or morphological novelties.